Description and short video of the alignment view functionality
In this article we will explain what the explorer view is and how it can be used.
This webpage allows you to slice and dice your search results to already determine certain answers to your research purpose.
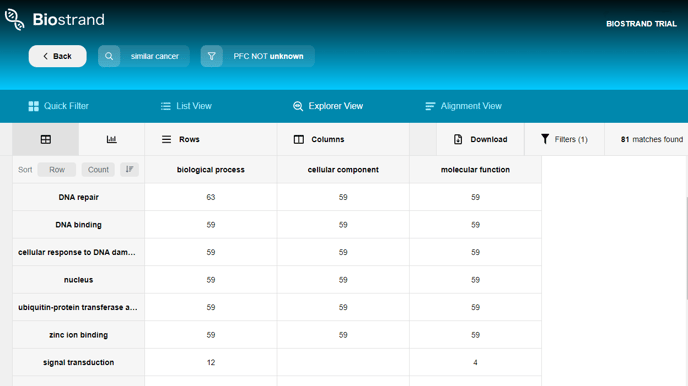
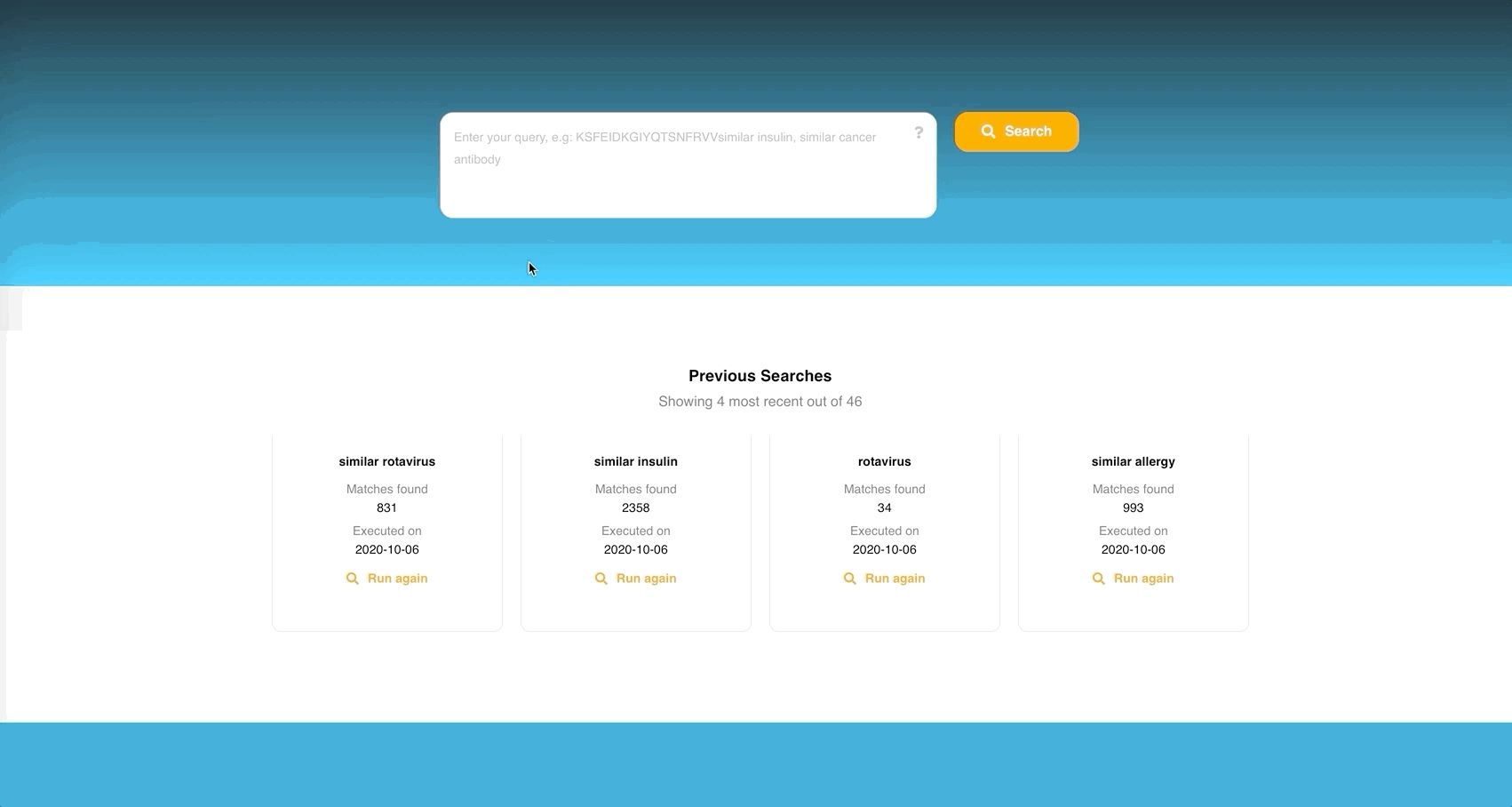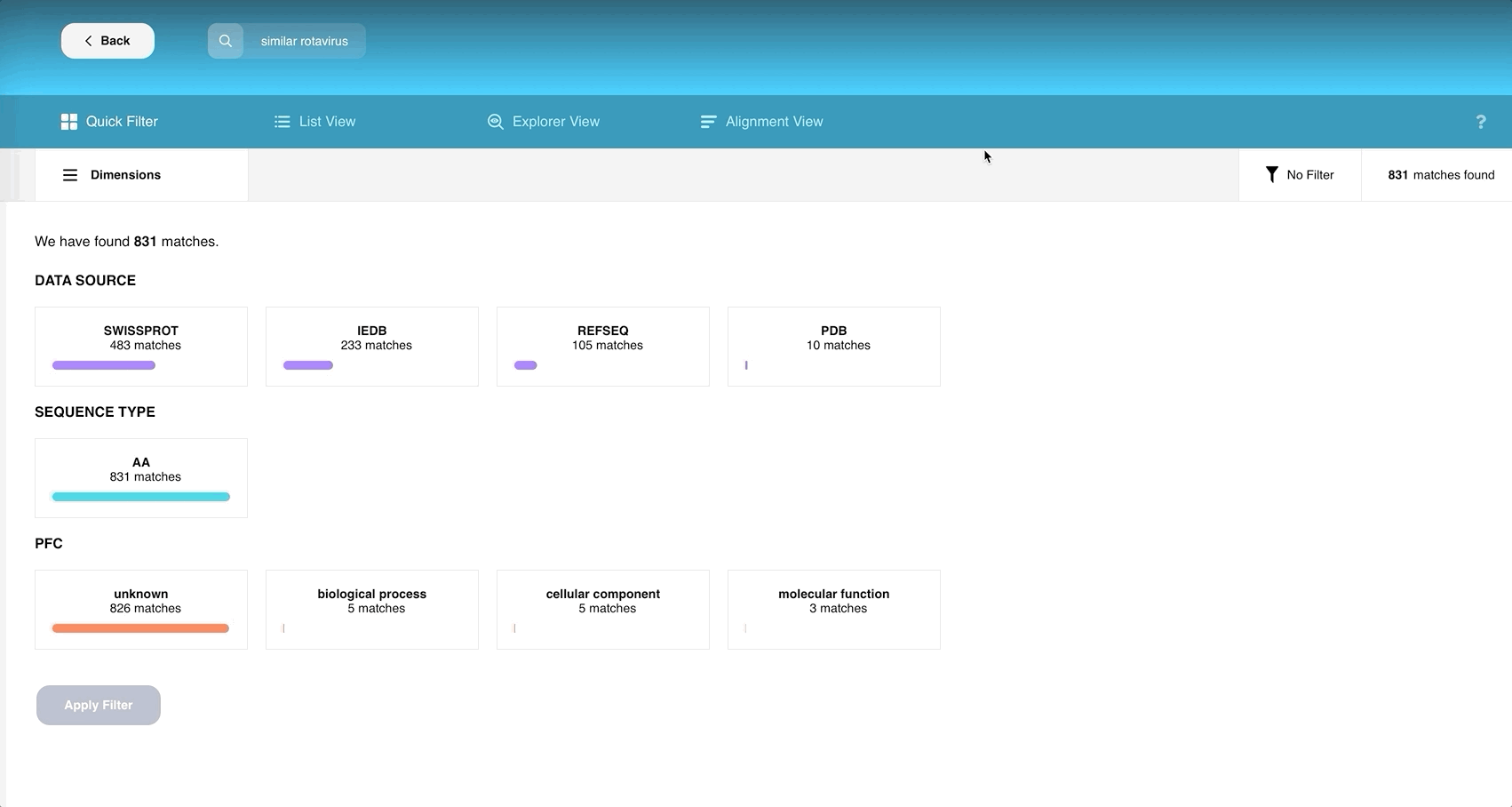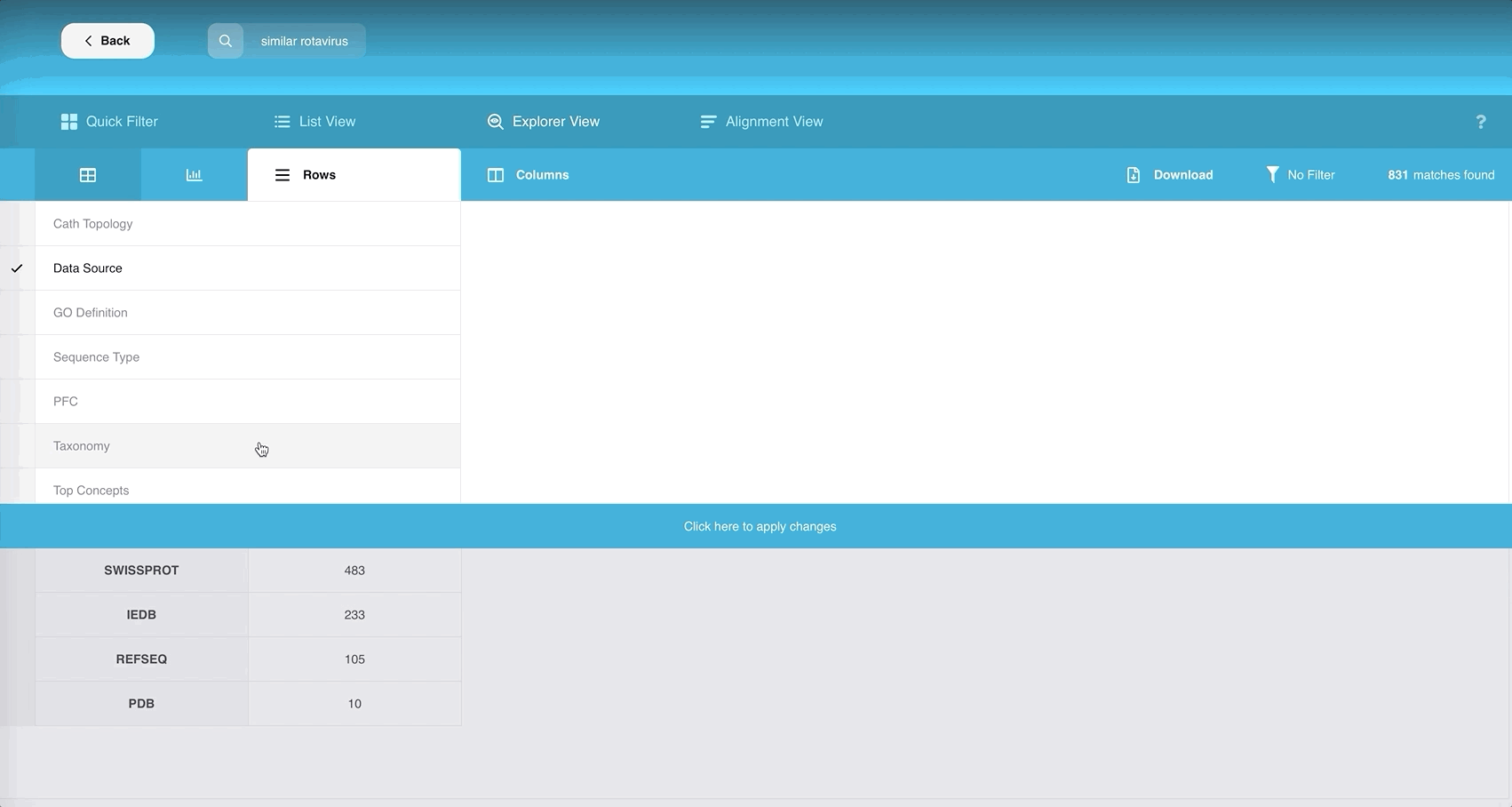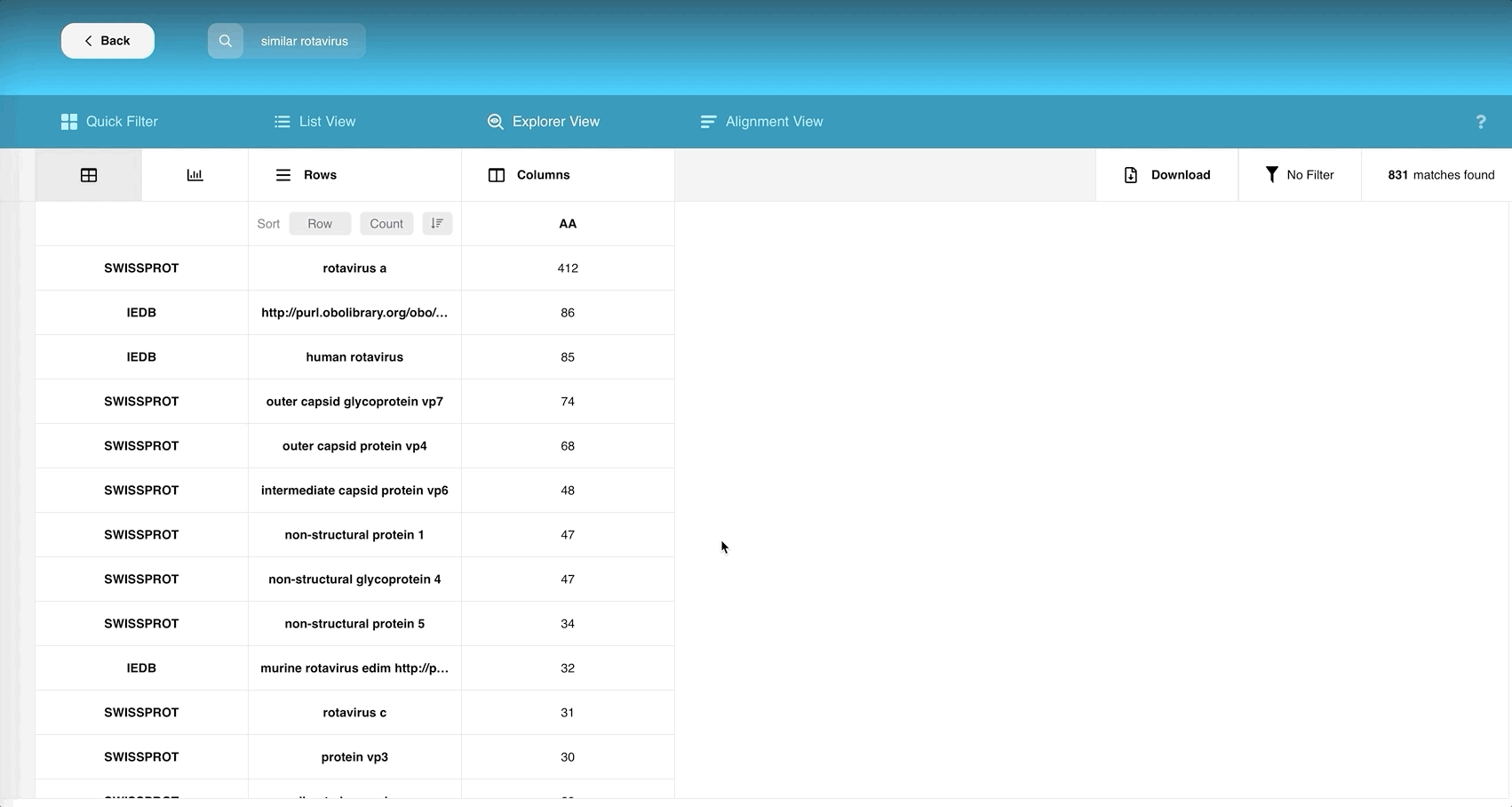
If we compare with other pages, the additional panel in this view contains the buttons “Rows”, “Columns”, “Download” and “No Filter”. The total number of search results after the latest filtering is displayed on the far right.
Both “Rows” and “Columns” currently allow you to choose at least one field and at most three fields respectively that can describe your table of records.
The list of domains to choose from is mostly the same as that described in the List View:
-
Data Source: Database which contains this information
-
Sequence Type: Either of DNA/RNA/AA
-
Gene Ontology (PFC): General definition of what aspects this sequence affects in the organism [biological process/molecular function/cellular component]
-
Taxonomy
-
Top concepts
You can choose multiple rows and multiple columns to find the common data points among the chosen attributes.
Remember to click “Apply Changes”